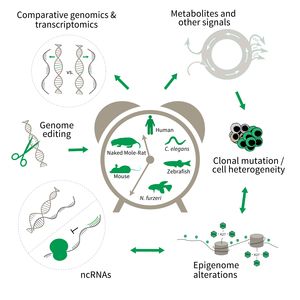
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2016
- Analysis of the oncogenic potential of the Wilms tumor suppressor gene 1 (Wt1)
Reichardt C
Dissertation 2016, Jena, Germany - Outgroups and Positive Selection: The Nothobranchius furzeri Case.
Sahm A, Platzer M, Cellerino A
Trends Genet 2016, 32(9), 523-5 - MHC-dependent mate choice is linked to a trace-amine-associated receptor gene in a mammal.
Santos PSC, Courtiol A, Heidel AJ, Höner OP, Heckmann I, Nagy M, Mayer F, Platzer M, Voigt CC, Sommer S
Sci Rep 2016, 6, 38490 - Genetic Factors of the Disease Course after Sepsis: A Genome-Wide Study for 28Day Mortality.
Scherag A, Schöneweck F, Kesselmeier M, Taudien S, Platzer M, Felder M, Sponholz C, Rautanen A, Hill AVS, Hinds CJ, Hossain H, Suttorp N, Kurzai O, Slevogt H, Giamarellos-Bourboulis EJ, Armaganidis A, Trips E, Scholz M, Brunkhorst FM
EBioMedicine 2016, 12, 239-46 - Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals.
Schwörer S, Becker F, Feller C, Baig AH, Köber U, Henze H, Kraus JM, Xin B, Lechel A, Lipka DB, Varghese CS, Schmidt M, Rohs R, Aebersold R, Medina KL, Kestler HA, Neri F, von Maltzahn** J, Tümpel** S, Rudolph** KL
Nature 2016, 540(7633), 428-32 ** co-corresponding authors - Polymorphisms of cystathionine beta-synthase gene are associated with susceptibility to sepsis.
Sponholz C, Kramer M, Schöneweck F, Menzel U, Inanloo Rahatloo K, Giamarellos-Bourboulis EJ, Papavassileiou V, Lymberopoulou K, Pavlaki M, Koutelidakis I, Perdios I, Scherag A, Bauer M, Platzer M, Huse K
Eur J Hum Genet 2016, 24(7), 1041-8 - Genetic Factors of the Disease Course After Sepsis: Rare Deleterious Variants Are Predictive.
Taudien* S, Lausser* L, Giamarellos-Bourboulis EJ, Sponholz C, Schöneweck F, Felder M, Schirra LR, Schmid F, Gogos C, Groth S, Petersen BS, Franke A, Lieb W, Huse K, Zipfel PF, Kurzai O, Moepps B, Gierschik P, Bauer M, Scherag A, Kestler** HA, Platzer** M
EBioMedicine 2016, 12, 227-38 * equal contribution, ** co-senior authors - Finding approximate gene clusters with Gecko 3.
Winter S, Jahn K, Wehner S, Kuchenbecker L, Marz M, Stoye J, Böcker S
Nucleic Acids Res 2016, 44(20), 9600-10