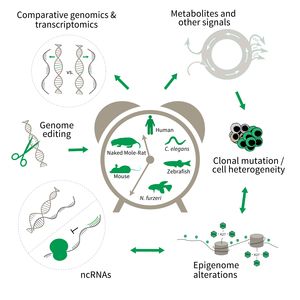
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2024
- Epigenetic clock and lifespan prediction in the short-lived killifish Nothobranchius furzeri
Giannuzzi C, Baumgart M, Neri F, Cellerino A
bioRxiv 2024, https://doi.org/10.1101/2024.08. - Immunity and bacterial recruitment in plant leaves are parallel processes whose link shapes sensitivity to temperature stress
Jose J, Teutloff E, Naseem S, Barth E, Halitschke R, Marz M, T.Agler M
bioRxiv 2024, https://doi.org/10.1101/2024.06. - Comprehensive transcriptome analysis reveals altered mRNA splicing and post-transcriptional changes in the aged mouse brain.
Kumar NH, Kluever V, Barth E, Krautwurst S, Furlan M, Pelizzola M, Marz M, Fornasiero EF
Nucleic Acids Res 2024, 52(6), 2865-85 - Single-cell sequencing analysis unravels genes/pathways regulating vertebrate aging
Meng Y
Dissertation 2024, Jena, Germany - Data report on gene expression after hepatic portal vein ligation (PVL) in rats.
Meyer** D, Kosacka J, von Bergen M, Christ B, Marz** M
Front Genet 2024, 15, 1421955 ** co-corresponding authors - Exploring the impact of primer length on efficient gene detection via high-throughput sequencing.
Micheel J, Safrastyan A, Aron F, Wollny D
Nat Commun 2024, 15(1), 5858 - Polyamines sustain epithelial regeneration in aged intestines by modulating protein homeostasis
Minetti A, Omrani O, Brenner C, Allies G, Imada S, Rösler J, Khawaled S, Cansiz F, W.Meckelmann S, Gebert N, Heinze I, Lu J, Spengler K, Rasa M, Heller R, Yilmaz O, Tasdogan A, Neri F, Ori A
bioRxiv 2024, https://doi.org/10.1101/2024.07. - Sex chromosome turnover in African annual killifishes of the genus Nothobranchius
Monika H, Pablo M, Anna CV, Ahmed AR, Sergey AS, Tomáš P, Marie A, Karolína J, Jana Š, Nikolas T, Marek J, Matyáš H, Liehr T, Martin R, Eugene YK, Petr R, Englert C, Petr N, Sember A
bioRxiv 2024, https://doi.org/10.1101/2024.03. - Surface microlayer-mediated virome dissemination in the Central Arctic.
Rahlff J, Westmeijer G, Weissenbach J, Antson A, Holmfeldt K
Microbiome 2024, 12(1), 218 - Viral challenges and adaptations between Central Arctic Ocean and atmosphere
Rahlff J, Westmeijer G, Weissenbach J, Antson A, Holmfeldt K
bioRxiv 2024, https://doi.org/10.1101/2024.03.