Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
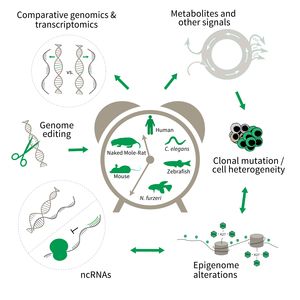
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2025
- Analysis of microRNA expression reveals convergent evolution of the molecular control of diapause in annual killifishes.
Barth* E, Baumgart* M, Dolfi* L, Cui R, Groth M, Ripa R, Savino A, Valenzano DR, Platzer M, Marz** M, Cellerino** A
Front Genet 2025, 16, 1583989 * equal contribution, ** co-corresponding authors - Translational Remodeling of the Synaptic Proteome During Aging.
Caterino C, Ugolini M, Durso W, Jevdokimenko K, Groth M, Riege K, Görlach M, Fornasiero E, Ori A, Hoffmann S, Cellerino A
Aging Cell 2025 (epub ahead of print) - Nerve Growth Factor Receptor (NGFR/p75NTR) of the Small-Spotted Catshark (Scyliorhinus canicula): Evolutionary Conservation and Brain Function.
Chiavacci E, Camera R, Costa M, Fronte B, Tozzini ET, Cellerino A
J Comp Neurol 2025, 533(4), e70049 - Altered translation elongation contributes to key hallmarks of aging in the killifish brain.
Di Fraia* D, Marino* A, Lee* JH, Kelmer Sacramento E, Baumgart M, Bagnoli S, Balla T, Schalk F, Kamrad S, Guan R, Caterino C, Giannuzzi C, Tomaz da Silva P, Sahu AK, Gut H, Siano G, Tiessen M, Terzibasi-Tozzini E, Fornasiero EF, Gagneur J, Englert C, Patil KR, Correia-Melo C, Nedialkova DD, Frydman** J, Cellerino** A, Ori** A
Science 2025, 389(6759), eadk3079 * equal contribution, ** co-senior authors - Wilms' tumor 1 impairs apoptotic clearance of fibroblasts in distal fibrotic lung lesions.
Ediga HH, Vemulapalli CP, Sontake V, Patel PK, Miyazaki H, Popov D, Jensen MB, Jegga AG, Huang SK, Englert C, Schedl A, Gupta N, McCormack FX, Madala SK
J Clin Invest 2025, 135(15), e188819 - AnchoRNA: Full virus genome alignments through conserved anchor regions
Eulenfeld** T, Triebel S, Marz** M
bioRxiv 2025, https://doi.org/10.1101/2025.01. ** co-corresponding authors - Multiple Origins of Sex Chromosomes in Nothobranchius Killifishes.
Hospodářská M, Mora P, Voleníková AC, Al-Rikabi A, Altmanová M, Simanovsky SA, Tolar N, Pavlica T, Janečková K, Štundlová J, Bobryshava K, Jankásek M, Hiřman M, Liehr T, Reichard M, Krysanov EY, Ráb P, Englert C, Nguyen P, Sember A
Mol Ecol 2025, 34(16), e70029 - Analysis of different strains of the turquoise killifish identify transcriptomic signatures associated with heritable lifespan differences.
Mazzetto M, Reichwald K, Koch P, Groth M, Cellerino A
J Gerontol A Biol Sci Med Sci 2025, 80(7), glae255 - diffONT: predicting methylation-specific PCR biomarkers based on nanopore sequencing data for clinical application
Meyer** D, Barth E, Wiehle L, Marz** M
bioRxiv 2025, https://doi.org/10.1101/2025.02. ** co-corresponding authors - Multiple Wnt signaling pathways direct epithelial tubule interconnection in the regenerating zebrafish kidney
N.Kamei C, G.B.Sampson W, Albertz C, Aries O, Wolf A, M.Upadhyay R, M.Hughes S, Schenk H, Bonnet F, W.Draper B, W.McCracken K, K.Marciano D, Oxburgh L, A.Drummond I
bioRxiv 2025, https://doi.org/10.1101/2025.03.