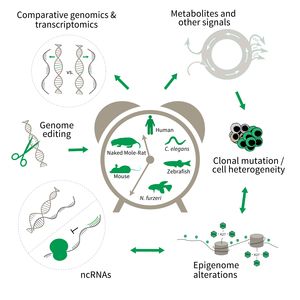
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2016
- Semantic multi-classifier systems for the analysis of gene expression profiles
Lausser* L, Schmid* F, Platzer M, Sillanpää JM, Kestler AH
Data Science Series A (Online-First) 2016, 1(1) * equal contribution - Impaired Planar Germ Cell Division in the Testis, Caused by Dissociation of RHAMM from the Spindle, Results in Hypofertility and Seminoma.
Li H, Frappart* L, Moll* J, Winkler* A, Kroll T, Hamann J, Kufferath I, Groth M, Taudien S, Schütte M, Yaspo ML, Heuer H, Lange BMH, Platzer M, Zatloukal K, Herrlich P, Ploubidou A
Cancer Res 2016, 76(21), 6382-95 * equal contribution - Dicer ablation in osteoblasts by Runx2 driven cre-loxP recombination affects bone integrity, but not glucocorticoid-induced suppression of bone formation.
Liu P, Baumgart M, Groth M, Wittmann J, Jäck HM, Platzer M, Tuckermann* JP, Baschant* U
Sci Rep 2016, 6, 32112 * equal contribution - The distribution of α-kleisin during meiosis in the holocentromeric plant Luzula elegans.
Ma W, Schubert V, Martis MM, Hause G, Liu Z, Shen Y, Conrad U, Shi W, Scholz U, Taudien S, Cheng Z, Houben A
Chromosome Res 2016, 24(3), 393-405 - Coronavirus cis-Acting RNA Elements.
Madhugiri R, Fricke M, Marz M, Ziebuhr J
In: Advances in Virus Research (edited by John Ziebuhr) 2016, 96, 127-163, Elsevier Inc. Academic - The long intergenic non-coding RNA CCR492 functions as a let-7 competitive endogenous RNA to regulate c-Myc expression.
Maldotti M, Incarnato D, Neri F, Krepelova A, Rapelli S, Anselmi F, Parlato C, Basile G, Dettori D, Calogero R, Oliviero S
BBA-GENE REGUL MECH 2016, 1859(10), 1322-32 published during change of institution - Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq.
Marthandan S, Baumgart M, Priebe S, Groth M, Schaer J, Kaether C, Guthke R, Cellerino A, Platzer M, Diekmann S, Hemmerich P
PLoS One 2016, 11(5), e0154531 - Conserved genes and pathways in primary human fibroblast strains undergoing replicative and radiation induced senescence.
Marthandan S, Menzel U, Priebe S, Groth M, Guthke R, Platzer M, Hemmerich P, Kaether C, Diekmann S
Biol Res 2016, 49(1), 34 - Olfactory phenotypic expression unveils human aging.
Mazzatenta A, Cellerino A, Origlia N, Barloscio D, Sartucci F, Di Giulio C, Domenici L
Oncotarget 2016, 7(15), 19193-200 - Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis.
Neri F, Incarnato D, Krepelova A, Parlato C, Oliviero S
Nat Protoc 2016, 11(7), 1191-205 published during change of institution