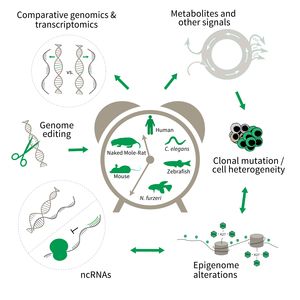
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2025
- Nerve Growth Factor Receptor (NGFR/p75NTR) of the Small-Spotted Catshark (Scyliorhinus canicula): Evolutionary Conservation and Brain Function.
Chiavacci E, Camera R, Costa M, Fronte B, Tozzini ET, Cellerino A
J Comp Neurol 2025, 533(4), e70049 - AnchoRNA: Full virus genome alignments through conserved anchor regions
Eulenfeld** T, Triebel S, Marz** M
bioRxiv 2025, https://doi.org/10.1101/2025.01. ** co-corresponding authors - Analysis of different strains of the turquoise killifish identify transcriptomic signatures associated with heritable lifespan differences.
Mazzetto M, Reichwald K, Koch P, Groth M, Cellerino A
J Gerontol A Biol Sci Med Sci 2025 (epub ahead of print) - The master male sex determinant Gdf6Y of the turquoise killifish arose through allelic neofunctionalization.
Richter A, Mörl H, Thielemann M, Kleemann M, Geißen R, Schwarz R, Albertz C, Koch P, Petzold A, Kroll T, Groth M, Hartmann N, Herpin A, Englert C
Nat Commun 2025, 16(1), 540
2024
- Quantitative mapping of proteasome interactomes and substrates using ProteasomeID.
Bartolome A, Heiby* JC, Di Fraia* D, Heinze I, Knaudt H, Spaeth E, Omrani O, Minetti A, Hofmann M, Kirkpatrick JM, Dau** T, Ori** A
Elife 2024, 13, RP93256 * equal contribution, ** co-corresponding authors - [Genetics, epigenetics, and environmental factors in life expectancy-What role does nature-versus-nurture play in aging?].
Bierhoff H
Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2024, 67(5), 521-7 - NRF2 activation by cysteine as a survival mechanism for triple-negative breast cancer cells.
Bottoni L, Minetti A, Realini G, Pio E, Giustarini D, Rossi R, Rocchio C, Franci L, Salvini L, Catona O, D'Aurizio R, Rasa M, Giurisato E, Neri F, Orlandini M, Chiariello M, Galvagni F
Oncogene 2024, 43(22), 1701-13 - The Aging Synapse: An Integrated Proteomic And Transcriptomic Analysis
Caterino* C, Ugolini* M, Durso W, Jevdokimenko K, Groth M, Riege K, Görlach M, Fornasiero E, Ori A, Hoffmann S, Cellerino A
bioRxiv 2024, https://doi.org/10.1101/2024.12. * equal contribution - Spatio-temporal variability of airborne viruses over sub-Antarctic South Georgia: Effects of sampling device and proximity of the ocean
Das R, Malard L, Pearce DA, Rahlff J
bioRxiv 2024, doi.org/10.1101/2024.08.23.60932 - Amyloid beta precursor protein contributes to brain aging and learning decline in short-lived turquoise killifish (Nothobranchius furzeri)
E.M.de Bakker D, Mihaljević M, Gharat K, Richter Y, Bagnoli S, van Bebber F, Adam L, Shamim-Schulze F, Ohlenschläger O, Bens M, Cirri E, Antebi A, Matić I, Schneider A, Schmid B, Cellerino A, Kirstein J, Riccardo Valenzano D
bioRxiv 2024, https://doi.org/10.1101/2024.10.