Subarea 1: Stem Cell Aging
The individual research groups within Subarea 1 investigate the causes and consequences of stem cell aging. The research work spans from basic model organisms over genetic mouse models up to humanized mouse models engrafted with human stem cells.
According to the FLI, with the closure of two groups since 2016 the representation of invertebrate models of stem cell research was reduced in Subarea 1. The institute presumes that the recruitment of new groups should fill this gap.
The research is defined by four focus areas:
- Cell-intrinsic mechanisms limiting the function of aging stem and progenitor cells,
- Aging-associated alterations of stem cell niches and the systemic environment,
- Mechanisms of clonal selection and epigenetic drifts in stem cell aging, and
- Microbiota- and metabolism-induced impairments in stem cell function during aging (in context of the new focus area Microbiota and Aging currently being built up within Subarea 2).
Research focus of Subarea 1.
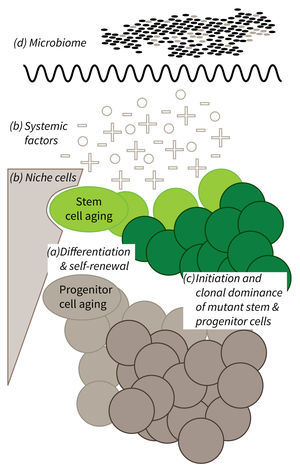
a) It is currently not well understood what mechanisms impair cellular functions in aging. b) The relative contribution of niche cells and systemic acting factors on stem cell aging have yet to be determined in different tissues. c) Clonal expansion of mutant cells associates with disease development in aging humans. Mechanistically, the process remains poorly understood. Changes in color intensity depict clonal dominance originating from stem (green) or progenitor cells (gray). d) Emerging evidences indicate that aging associated alter ations in microbiota influence stem cell function and vice versa.
Publications
(since 2016)
2024
- Denervation alters the secretome of myofibers and thereby affects muscle stem cell lineage progression and functionality.
Henze H, Hüttner SS, Koch P, Schüler SC, Groth M, von Eyss B, von Maltzahn J
NPJ Regen Med 2024, 9(1), 10 - The oncogenic lncRNA MIR503HG suppresses cellular senescence counteracting supraphysiological androgen treatment in prostate cancer.
Kallenbach J, Rasa M, Heidari Horestani M, Atri Roozbahani G, Schindler K, Baniahmad A
J Exp Clin Cancer Res 2024, 43(1), 321 - RelB and C/EBPα critically regulate the development of Peyer's patch mononuclear phagocytes.
Kanaya T, Jinnohara T, Sakakibara S, Tachibana N, Sasaki T, Kato T, Riemann M, Jin J, Shiroguchi K, Kawakami E, Ohno H
MUCOSAL IMMUNOL 2024 (epub ahead of print) - CRISPR/Cas9-Mediated Modification of PTP Expression.
Lossius C, Kresinsky A, Quiet L, Müller JP
Methods Mol Biol 2024, 2743, 43 - We need to talk-how muscle stem cells communicate.
Majchrzak K, Hentschel E, Hönzke K, Geithe C, von Maltzahn J
Front Cell Dev Biol 2024, 12, 1378548 - Optimizing Biocompatibility and Gene Delivery with DMAEA and DMAEAm: A Niacin-Derived Copolymer Approach.
Mapfumo PP, Reichel LS, André T, Hoeppener S, Rudolph LK, Traeger A
Biomacromolecules 2024, 25(8), 4749-61 - Vitamin B3 Containing Polymers for Nanodelivery.
Mapfumo PP, Solomun JI, Becker F, Moek E, Leiske MN, Rudolph LK, Brendel JC, Traeger A
Macromol Biosci 2024, e2400002 - Oncogenic Calreticulin Induces Immune Escape by Stimulating TGFβ Expression and Regulatory T-cell Expansion in the Bone Marrow Microenvironment.
Schmidt D, Endres C, Hoefflin R, Andrieux G, Zwick M, Karantzelis N, Staehle HF, Vinnakota JM, Duquesne S, Mozaffari Jovein M, Pfeifer D, Becker H, Blazar BR, Zähringer A, Duyster J, Brummer T, Boerries M, Baumeister J, Shoumariyeh K, Li J, Green AR, Heidel FH, Tirosh I, Pahl HL, Leimkühler N, Köhler N, de Toledo MAS, Koschmieder S, Zeiser R
Cancer Res 2024, 84(18), 2985-3003 - Consequences of GMPPB deficiency for neuromuscular development and maintenance.
Schurig MK, Umeh O, Henze* H, Jung* MJ, Gresing L, Blanchard V, von Maltzahn J, Hübner* CA, Franzka* P
Front Mol Neurosci 2024, 17, 1356326 * equal contribution - Pyjacker identifies enhancer hijacking events in acute myeloid leukemia including MNX1 activation via deletion 7q
Sollier E, Riedel A, H.Toprak U, A.Wierzbinska J, Weichenhan D, Philipp Schmid J, Hakobyan M, Touzart A, Jahn E, Vick B, Brown-Burke F, Kelly K, Kelekci S, Pejkovska A, Goyal A, Bähr M, Breuer K, May Chen MJ, Llamazares-Prada M, Hartmann M, Schönung M, Correia N, Trumpp A, Abdullah Y, Klingmüller U, S.Mughal S, Brors B, Westermann F, Schlesner M, Vosberg S, Herold T, A.Greif P, Pfeifer D, Lübbert M, Fischer T, H.Heidel F, Gebhard C, Walter W, Haferlach T, Eisfeld AK, Mrózek K, Nicolet D, Bullinger L, Smeenk L, Erpelinck C, Mulet-Lazaro R, Delwel R, Ernst A, Scherer M, Lutsik P, Jeremias I, Döhner K, Döhner H, B.Lipka D, Plass C
bioRxiv 2024, https://doi.org/10.1101/2024.09.