Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
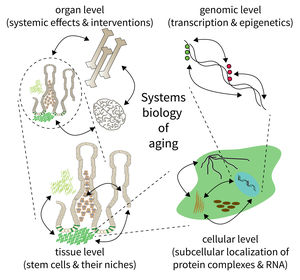
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2025
- Synergistic potential of CDK4/6 inhibitors and ATRA in non-APL AML.
Skopek R, Özcan SG, Chmiel P, Morgner S, Schütt J, Ghazvini Zadegan F, Stanko C, Palusińska M, Maślińska-Gromadka K, Sbirkov Y, Stengel S, Fischer M, Brioli A, Zelent A, Szymański Ł, Schenk T
Br J Haematol 2025 (epub ahead of print) - UV laser crosslinking uncovers novel DNA-binding pattern of CTCF*
Stanko C, Gupse Özcan S, Stengel S, Szymański Ł, Steube A, Fischer M, Brioli A, Schenk T
bioRxiv 2025, https://doi.org/10.1101/2025.05. - Npbwr1 signaling mediates fast antidepressant action.
Stein G, Aly JS, Lange L, Manzolillo A, Riege K, Brancato A, Hübner CA, Turecki G, Hoffmann S, Engmann O
Mol Psychiatry 2025, 30(5), 1828-35 - Age-dependent gut microbiota dynamics and their association with male fitness traits in Drosophila melanogaster
Sultanova** * Z, Dönertaş** * HM, Hita A, Aguilar P, Dag B, Ignacio Lucas-Lledo J, Latorre A, Carazo P
bioRxiv 2025, https://doi.org/10.1101/2025.05. * equal contribution, ** co-senior authors - Dynamics of HBV biomarkers during nucleos(t)ide analog treatment: A 14-year study.
van Bömmel F, Degasperi E, van Bömmel A, Facchetti F, Sambarino D, Deichsel D, Brehm J, Kamga Wouambo R, Maier M, Pfefferkorn M, Berg T, Lampertico P
Hepatol Commun 2025, 9(6), e0708 - Author Correction: Large-scale benchmarking of circRNA detection tools reveals large differences in sensitivity but not in precision.
Vromman M, Anckaert J, Bortoluzzi S, Buratin A, Chen CY, Chu Q, Chuang TJ, Dehghannasiri R, Dieterich C, Dong X, Flicek P, Gaffo E, Gu W, He C, Hoffmann S, Izuogu O, Jackson MS, Jakobi T, Lai EC, Nuytens J, Salzman J, Santibanez-Koref M, Stadler P, Thas O, Vanden Eynde E, Verniers K, Wen G, Westholm J, Yang L, Ye CY, Yigit N, Yuan GH, Zhang J, Zhao F, Vandesompele J, Volders PJ
Nat Methods 2025, 22(2), 448 - Gene regulation by convergent promoters.
Wiechens E, Vigliotti* F, Siniuk* K, Schwarz R, Schwab K, Riege K, van Bömmel A, Görlich I, Bens M, Sahm A, Groth M, Sammons MA, Loewer A, Hoffmann** S, Fischer** M
Nat Genet 2025, 57(1), 206-17 * equal contribution, ** co-corresponding authors
2024
- INHBA is enriched in HPV-negative oropharyngeal squamous cell carcinoma and promotes cancer progression.
Abou Kors T, Hofmann L, Betzler A, Payer K, Bens M, Truong J, von Witzleben A, Thomas J, Kraus JM, Kalaajieh R, Huber D, Ezić J, Benckendorff J, Greve J, Schuler PJ, Ottensmeier CH, Kestler HA, Hoffmann TK, Theodoraki MN, Brunner C, Laban S
Cancer Res Commun 2024, 4(2), 571-87 - Multi-omics analysis of overexpressed tumor-associated proteins: gene expression, immunopeptide presentation, and antibody response in oropharyngeal squamous cell carcinoma, with a focus on cancer-testis antigens.
Abou Kors T, Meier M, Mühlenbruch L, Betzler AC, Oliveri F, Bens M, Thomas J, Kraus JM, Doescher J, von Witzleben A, Hofmann L, Ezic J, Huber D, Benckendorff J, Barth TFE, Greve J, Schuler PJ, Brunner C, Blackburn JM, Hoffmann TK, Ottensmeier C, Kestler HA, Rammensee HG, Walz JS, Laban S
Front Immunol 2024, 15, 1408173 - Quantitative mapping of proteasome interactomes and substrates using ProteasomeID.
Bartolome A, Heiby* JC, Di Fraia* D, Heinze I, Knaudt H, Spaeth E, Omrani O, Minetti A, Hofmann M, Kirkpatrick JM, Dau** T, Ori** A
Elife 2024, 13, RP93256 * equal contribution, ** co-corresponding authors