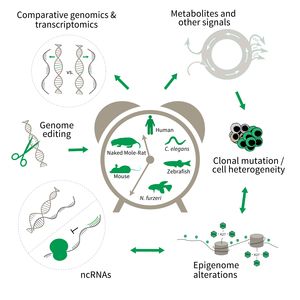
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2018
- Structural and functional conservation of cis-acting RNA elements in coronavirus 5'-terminal genome regions.
Madhugiri R, Karl N, Petersen D, Lamkiewicz K, Fricke M, Wend U, Scheuer R, Marz M, Ziebuhr J
Virology 2018, 517, 44-55 - The age-regulated zinc finger factor ZNF367 is a new modulator of neuroblast proliferation during embryonic neurogenesis.
Naef V, Monticelli S, Corsinovi D, Mazzetto MT, Cellerino A, Ori M
Sci Rep 2018, 8(1), 11836 - Dispersion/reaggregation in early development of annual killifishes: Phylogenetic distribution and evolutionary significance of a unique feature.
Naumann B, Englert C
Dev Biol 2018, 442(1), 69-79 - The impact of the Wilms tumor suppressor genes on zebrafish kidney regeneration.
Naumann U
Dissertation 2018, Jena, Germany - The Body’s Library
Neri F
GIT Laboratory Journal Europe 2018, 2, 16-9 - The high degree of cystathionine β-synthase (CBS) activation by S-adenosylmethionine (SAM) may explain naked mole-rat's distinct methionine metabolite profile compared to mouse.
Olecka M, Huse K, Platzer M
Geroscience 2018, 40(4), 359-60 - Exploring the basis of naked mole-rat healthspan by characterization of its transsulfuration pathway and hydroxymethylome.
Olecka MK
Dissertation 2018, Jena, Germany - Loss of Wilms tumor 1 protein is a marker for apoptosis in response to replicative stress in leukemic cells.
Pons* M, Reichardt* CM, Hennig D, Nathan A, Kiweler N, Stocking C, Wichmann C, Christmann M, Butter F, Reichardt S, Schneider G, Heinzel T, Englert C, Hartkamp J, Krämer OH, Mahendrarajah N
Arch Toxicol 2018, 92(6), 2119-35 * equal contribution - The Pseudogenes of Barley.
Prade VM, Gundlach H, Twardziok S, Chapman B, Tan C, Langridge P, Schulman AH, Stein N, Waugh R, Zhang G, Platzer M, Li C, Spannagl M, Mayer KFX
Plant J 2018, 93(3), 502-14 - Impairing L-Threonine Catabolism Promotes Healthspan through Methylglyoxal-Mediated Proteohormesis.
Ravichandran M, Priebe S, Grigolon G, Rozanov L, Groth M, Laube B, Guthke R, Platzer M, Zarse K, Ristow M
Cell Metab 2018, 27(4), 914-25