Hoffmann Research Group
Additional Projects
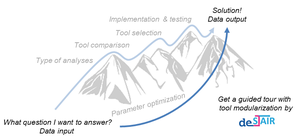
Joint Project „de.NBI – Partner: de.STAIR“
Bioinformatics Services for Structured Analysis and Integration of RNA-Seq experiments (de.STAIR)
Research focuses on RNA-Seq analysis, to develop best practices and user-friendly workflows for processing and integrating gene expression data as well as to provide assistance for experiment design, analysis and interpretation. The variety of RNA-Seq study designs and characteristic properties of studied organisms greatly affect bioinformatics analysis. As part of the RNA Bioinformatics center (RBC), which supports all RNA-related research within de.NBI, de.STAIR summarizes the expertise of three participating laboratories to develop and maintain an flexible RNA-Seq analysis workbench for different RNA-Seq protocols, to be used locally or in a cloud environment as provided by the open source Galaxy platform for biomedical research. In addition, de.STAIR provides workshops, training programs and screencasts for experts and non-experts to lower the barriers to RNA-Seq data analysis.
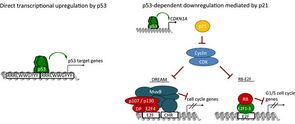
Mapping the indirect p53 gene regulatory network
Research focuses on dissecting gene regulatory networks that control the cell cycle and functions of the tumor suppressor p53. For example, the transcription factor p53 serves as a central suppressor of tumor progression. It controls cell proliferation and apoptosis by regulating a plethora of target genes. However, it is not clear how p53 regulates many of its target genes, what factors besides p53 itself are necessary for up or downregulation and what their regulation contributes to a normal or cancer cell. Our incomplete picture of the molecular basis of p53-dependent gene regulation remains a critical gap to our overall understanding of tumor suppression.
Contact

Steve Hoffmann
Group Leader
+49 3641 656810
steve.hoffmann@~@leibniz-fli.de
Patricia Möckel
Secretary
+49 3641 65-6240
patricia.moeckel@~@leibniz-fli.de
Team*
| Name | Phone | Position | |
|---|---|---|---|
| Steve Hoffmann | +49 3641 656810 | Group Leader | |
| Martin Fischer | +49 3641 656876 | Staff Scientist | |
| Konstantin Riege | +49 3641 656875 | Staff Scientist | |
| Alena van Bömmel | +49 3641 656868 | Staff Scientist | |
| Maja Kinga Olecka | +49 3641 656877 | Postdoc | |
| Omid Omrani | --- | Postdoc | |
| Kanstantin Siniuk | --- | Postdoc | |
| Atakan Ayden | +49 3641 656874 | Doctoral Candidate | |
| Aleksandr Cherkasov | +49 3641 656876 | Doctoral Candidate | |
| Tushar Patel | +49 3641 656868 | Doctoral Candidate | |
| Elina Wiechens | +49 3641 656123 | Doctoral Candidate | |
| Katjana Schwab | +49 3641 656877 | Scientist | |
| Robert Schwarz | +49 3641 656057 | Scientist | |
| Silke Förste | +49 3641 656243 | Technical Assistant | |
| Sandra Fischer | --- | Master Student | |
| Lara Sophie Schneider | --- | Bachelor Student |
* incomplete due to Data protection