Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
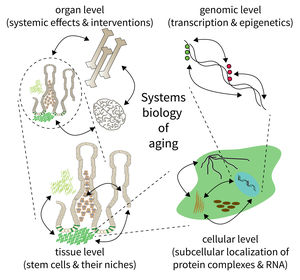
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2022
- LINC00892 Is an lncRNA Induced by T Cell Activation and Expressed by Follicular Lymphoma-Resident T Helper Cells.
Iaccarino I, Mourtada F, Reinke S, Patil P, Doose G, Monaco G, Hoffmann S, Siebert R, Klapper W
Noncoding RNA 2022, 8(3) - Inter-tissue convergence of gene expression during ageing suggests age-related loss of tissue and cellular identity.
Izgi H, Han D, Isildak U, Huang S, Kocabiyik E, Khaitovich** P, Somel** M, Dönertaş** HM
Elife 2022, 11, e68048 ** co-corresponding authors - Taz protects hematopoietic stem cells from an aging-dependent decrease in PU.1 activity.
Kim* KM, Mura-Meszaros* A, Tollot* M, Krishnan MS, Gründl M, Neubert L, Groth M, Rodriguez-Fraticelli A, Svendsen AF, Campaner S, Andreas N, Kamradt T, Hoffmann S, Camargo FD, Heidel FH, Bystrykh LV, de Haan G, von Eyss B
Nat Commun 2022, 13(1), 5187 * equal contribution - Protein lifetimes in aged brains reveal a proteostatic adaptation linking physiological aging to neurodegeneration.
Kluever V, Russo B, Mandad S, Kumar NH, Alevra M, Ori A, Rizzoli SO, Urlaub H, Schneider A, Fornasiero EF
Sci Adv 2022, 8(20), eabn4437 - Multifaceted Microcephaly-Related Gene MCPH1
Kristofova M, Ori A, Wang ZQ
Cells 2022, 11(2), 275 - CLN3 is required for the clearance of glycerophosphodiesters from lysosomes.
Laqtom NN, Dong W, Medoh UN, Cangelosi AL, Dharamdasani V, Chan SH, Kunchok T, Lewis CA, Heinze I, Tang R, Grimm C, Dang Do AN, Porter FD, Ori A, Sabatini DM, Abu-Remaileh M
Nature 2022, 609(7929), 1005-11 - Inflammaging is driven by upregulation of innate immune receptors and systemic interferon signaling and is ameliorated by dietary restriction.
Rasa* SMM, Annunziata* F, Krepelova A, Nunna S, Omrani O, Gebert N, Adam L, Käppel S, Höhn S, Donati G, Jurkowski TP, Rudolph KL, Ori A, Neri F
Cell Rep 2022, 39(13), 111017 * equal contribution - PLCG1 is required for AML1-ETO leukemia stem cell self-renewal.
Schnoeder TM, Schwarzer A, Jayavelu AK, Hsu CJ, Kirkpatrick J, Döhner K, Perner F, Eifert T, Huber N, Arreba-Tutusaus P, Dolnik A, Assi SA, Nafria M, Jiang L, Dai YT, Chen Z, Chen SJ, Kellaway SG, Ptasinska A, Ng ES, Stanley EG, Elefanty AG, Buschbeck M, Bierhoff H, Brodt S, Matziolis G, Fischer KD, Hochhaus A, Chen CW, Heidenreich O, Mann M, Lane SW, Bullinger L, Ori A, Eyss Bv, Bonifer C, Heidel F
Blood 2022, 139(7), 1080-97 - Locus-specific expression analysis of transposable elements.
Schwarz R, Koch P, Wilbrandt* J, Hoffmann* S
Brief Bioinform 2022, 23(1), bbab417 * equal contribution - Single-cell atlas of the aging mouse colon.
Širvinskas D, Omrani O, Lu J, Rasa M, Krepelova A, Adam L, Kaeppel S, Sommer F, Neri F
iScience 2022, 25(5), 104202