Subarea 4: Cell Dynamics and Molecular Damages in Aging
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
Research focus of Subarea 4.
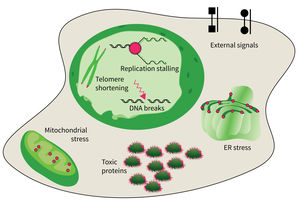
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2020
- Characterization of two Caenorhabditis elegans Orthologs of Selenium-binding protein 1, associated with aging
Köhnlein K
Dissertation 2020, Jena, Germany - A Caenorhabditis elegans ortholog of human selenium-binding protein 1 is a pro-aging factor protecting against selenite toxicity.
Köhnlein* K, Urban* N, Guerrero-Gómez D, Steinbrenner H, Urbánek P, Priebs J, Koch P, Kaether C, Miranda-Vizuete A, Klotz LO
Redox Biol 2020, 28, 101323 * equal contribution - Biomimetic reconstruction of the hematopoietic stem cell niche for in vitro amplification of human hematopoietic stem cells.
Marx-Blümel L, Marx C, Weise F, Frey J, Perner B, Schlingloff G, Lindig N, Hampl J, Sonnemann J, Brauer D, Voigt A, Singh S, Beck B, Jäger UM, Wang ZQ, Beck JF, Schober A
PLoS One 2020, 15(6), e0234638 - Systematic elucidation of neuron-astrocyte interaction in models of amyotrophic lateral sclerosis using multi-modal integrated bioinformatics workflow.
Mishra V, Re DB, Le Verche V, Alvarez MJ, Vasciaveo A, Jacquier A, Doulias PT, Greco TM, Nizzardo M, Papadimitriou D, Nagata T, Rinchetti P, Perez-Torres EJ, Politi KA, Ikiz B, Clare K, Than ME, Corti S, Ischiropoulos H, Lotti F, Califano A, Przedborski S
Nat Commun 2020, 11(1), 5579 - Growth inhibitory role of the p53 activator SCH 529074 in non‑small cell lung cancer cells expressing mutant p53.
Nenkov M, Ma Y, Haase D, Zhou Z, Li Y, Petersen I, Lu G, Chen Y
Oncol Rep 2020, 43(2), 2073-82 - Tissue-specific Gene Expression Changes Are Associated with Aging in Mice.
Srivastava* A, Barth* E, Ermolaeva MA, Guenther M, Frahm C, Marz** M, Witte** OW
Genomics Proteomics Bioinformatics 2020, 18(4), 430-42 * equal contribution, ** co-corresponding authors - NBS1 interacts with Notch signaling in neuronal homeostasis.
Zhou ZW, Kirtay M, Schneble N, Yakoub G, Ding M, Rüdiger T, Siniuk K, Lu R, Jiang YN, Li TL, Kaether C, Barzilai A, Wang ZQ
Nucleic Acids Res 2020, 48(19), 10924-39
2019
- C/EBPβ-LIP induces cancer-type metabolic reprogramming by regulating the let-7 /LIN28B circuit in mice.
Ackermann T, Hartleben* G, Müller* C, Mastrobuoni G, Groth M, Sterken BA, Zaini MA, Youssef SA, Zuidhof HR, Krauss SR, Kortman G, de Haan G, de Bruin A, Wang ZQ, Platzer M, Kempa S, Calkhoven CF
Commun Biol 2019, 2, 208 * equal contribution - In vivo Study of Human DHX9 Helicase Functions
Aly YSH
Dissertation 2019, Jena, Germany - A disease causing ATLASTIN 3 mutation affects multiple endoplasmic reticulum-related pathways.
Behrendt L, Kurth I, Kaether C
Cell Mol Life Sci 2019, 76(7), 1433-45