Subarea 4: Cell Dynamics and Molecular Damages in Aging
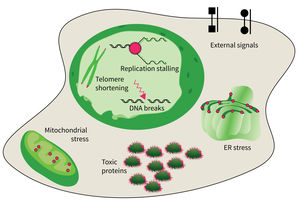
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
Research focus of Subarea 4.
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2021
- Poly(ADP-ribose) regulation in cell cycle control and cancer therapy
Siniuk K
Dissertation 2021, Jena, Germany - HAT cofactor TRRAP modulates microtubule dynamics via SP1 signaling to prevent neurodegeneration.
Tapias* A, Lázaro* D, Yin* BK, Rasa SMM, Krepelova A, Kelmer Sacramento E, Grigaravicius P, Koch P, Kirkpatrick J, Ori A, Neri F, Wang ZQ
Elife 2021, 10, e61531 * equal contribution - Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription.
Yin BK, Wang ZQ
Int J Mol Sci 2021, 22(22)
2020
- Bring it back, bring it back, don't take it away from me - the sorting receptor RER1.
Annaert W, Kaether C
J Cell Sci 2020, 133(17) - Atlastine – wie defekte Netzwerke Neuropathien verursachen
Behrendt L, Kaether C
BIOspektrum 2020, 5, 485-7 - Multiple biochemical properties of the p53 molecule contribute to activation of polymerase iota-dependent DNA damage tolerance.
Biber S, Pospiech H, Gottifredi V, Wiesmüller L
Nucleic Acids Res 2020, 48(21), 12188-203 - Loss of metabolic plasticity underlies metformin toxicity in aged Caenorhabditis elegans.
Espada* L, Dakhovnik* A, Chaudhari* P, Martirosyan A, Miek L, Poliezhaieva T, Schaub Y, Nair A, Döring N, Rahnis N, Werz O, Koeberle A, Kirkpatrick J, Ori A, Ermolaeva MA
Nat Metab 2020, 2(11), 1316-31 * equal contribution - Bi-allelic HPDL Variants Cause a Neurodegenerative Disease Ranging from Neonatal Encephalopathy to Adolescent-Onset Spastic Paraplegia.
Husain RA, Grimmel M, Wagner M, Hennings JC, Marx C, Feichtinger RG, Saadi A, Rostásy K, Radelfahr F, Bevot A, Döbler-Neumann M, Hartmann H, Colleaux L, Cordts I, Kobeleva X, Darvish H, Bakhtiari S, Kruer MC, Besse A, Ng ACH, Chiang D, Bolduc F, Tafakhori A, Mane S, Ghasemi Firouzabadi S, Huebner AK, Buchert R, Beck-Woedl S, Müller AJ, Laugwitz L, Nägele T, Wang ZQ, Strom TM, Sturm M, Meitinger T, Klockgether T, Riess O, Klopstock T, Brandl U, Hübner CA, Deschauer M, Mayr JA, Bonnen PE, Krägeloh-Mann I, Wortmann SB, Haack TB
Am J Hum Genet 2020, 107(2), 364-73 - Cell Metabolic Alterations due to Mcph1 Mutation in Microcephaly.
Journiac N, Gilabert-Juan J, Cipriani S, Benit P, Liu X, Jacquier S, Faivre V, Delahaye-Duriez A, Csaba Z, Hourcade T, Melinte E, Lebon S, Violle-Poirsier C, Oury JF, Adle-Biassette H, Wang ZQ, Mani S, Rustin P, Gressens P, Nardelli J
Cell Rep 2020, 31(2), 107506 - ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration.
Kidiyoor GR, Li Q, Bastianello G, Bruhn C, Giovannetti I, Mohamood A, Beznoussenko GV, Mironov A, Raab M, Piel M, Restuccia U, Matafora V, Bachi A, Barozzi S, Parazzoli D, Frittoli E, Palamidessi A, Panciera T, Piccolo S, Scita G, Maiuri P, Havas KM, Zhou ZW, Kumar A, Bartek J, Wang ZQ, Foiani M
Nat Commun 2020, 11(1), 4828