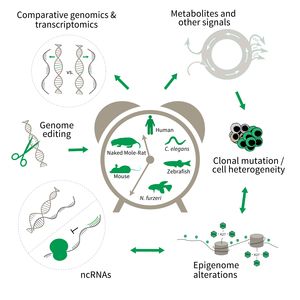
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2023
- Adapting the pantograph limb: Differential robustness of fore- and hindlimb kinematics against genetically induced perturbation in the neural control networks and its evolutionary implications.
Schnerwitzki D, Englert C, Schmidt M
Zoology (Jena) 2023, 157, 126076 - Tau alters global gene expression affecting chromatin in the early stages of Alzheimer’s disease
Siano* G, Varisco* M, Terrigno M, Wang C, Groth M, Galas MC, Hoozemans JJM, Cellerino A, Cattaneo** A, Di Primio** C
bioRxiv 2023, 10.1101/2023.11.17.567548 * equal contribution, ** co-senior authors - Magnipore: Prediction of differential single nucleotide changes in the Oxford Nanopore Technologies sequencing signal of SARS-CoV-2 samples
Spangenberg J, Höner zu Siederdissen C, Žarković M, Triebel S, Rose R, Martínez Christophersen C, Paltzow L, M.Hegab M, Wansorra A, Srivastava A, Krumbholz** A, Marz** M
bioRxiv 2023, https://doi.org/10.1101/2023.03. ** co-senior authors - Comprehensive Survey of Conserved RNA Secondary Structures in Full-Genome Alignment of Hepatitis C Virus
Triebel S, Lamkiewicz K, Ontiveros N, Sweeney B, Stadler PF, Petrov AI, Niepmann M, Marz M
bioRxiv 2023, 10.1101/2023.11.15.567179 - De novo genome assembly resolving repetitive structures enables genomic analysis of 35 European Mycoplasmopsis bovis strains.
Triebel S, Sachse K, Weber M, Heller M, Diezel C, Hölzer M, Schnee C, Marz M
BMC Genomics 2023, 24(1), 548 - De novo genome assembly resolving repetitive structures enables genomic analysis of 35 European Mycoplasma bovis strains
Triebel S, Sachse K, Weber M, Heller M, Diezel C, Hölzer M, Schnee C, Marz M
bioRxiv 2023, https://doi.org/10.1101/2023.04. - Characterization of myc mutants in the short-lived killifish Nothobranchius furzeri
Zhang Z
Dissertation 2023, Jena, Germany
2022
- Paneth cells drive intestinal stem cell competition and clonality in aging and calorie restriction
Annunziata F, Rasa SMM, Krepelova A, Lu J, Minetti A, Omrani O, Nunna S, Adam L, Käppel S, Neri F
Eur J Cell Biol 2022, 101(4), 151282 - Quantification of noradrenergic-, dopaminergic-, and tectal-neurons during aging in the short-lived killifish Nothobranchius furzeri.
Bagnoli S, Fronte B, Bibbiani C, Terzibasi Tozzini E, Cellerino A
Aging Cell 2022, 21(9), e13689 - The natural compound atraric acid suppresses androgen-regulated neo-angiogenesis of castration-resistant prostate cancer through angiopoietin 2.
Ehsani M, Bartsch S, Rasa SMM, Dittmann J, Pungsrinont T, Neubert L, Huettner SS, Kotolloshi R, Schindler K, Ahmad A, Mosig AS, Adam L, Ori A, Neri F, Berndt A, Grimm MO, Baniahmad A
Oncogene 2022, 41(23), 3263-77